Hauptinhalt
Topinformationen
Identification of novel genomic loci contributing to affective disorders
Complex multidimensional diseases such as major depression are determined by a variety of genetic and environmental factors, each of them ultimately contributing with minor effect size to the characteristic endophenotypes and the aetiology of the disease. The identification of genetic loci involved has therefore turned out to be complicated and was accompanied by setbacks. Genome-wide association studies (GWAS) constitute a controversial but so far irreplaceable tool for the identification of single loci involved in the genetic predisposition to develop complex diseases, including psychiatric disorders.
Despite growing scientific efforts, the genetic underpinnings of major depression and comorbid diseases such as generalized anxiety are still largely unknown. Therefore, we performed a GWAS using outbred CD-1 mice, i.e. the founder strain of the HR/IR/LR mouse model of affective disorders, with the aim of elucidating loci involved in the genetic determination of stress responsiveness, depression-like and anxiety-related behavioural phenotypes.
A total of 384 male CD-1 mice (treated in eight separate cohorts) were phenotypically characterised in a battery of tests assessing their emotional behaviour and the reactivity of the HPA axis. From these animals, 32 individuals were selected (representing the continuum of the respective quantitative traits) for a genome-wide high-throughput screening using the JAX Mouse Diversity Genotyping Array. This array simultaneously detects about 620,000 single nucleotide polymorphisms (SNPs) and about 916,000 copy number variants (CNVs) across the genome. The resulting pool of SNPs (a total of about 170,000 heterozygous or opposite homozygous SNPs were found) was used for the calculation of phenotype-genotype associations and for generating a first version of a CD-1 strain-specific haplotype map. Such a haplotype map allows the genome-wide selection of tag-SNPs in accordance with the linkage disequilibrium clustering of CD-1 mice and can serve as the basis for a future development of a customized tag-SNP assay testing associations in the entire CD-1 mouse population.
Our analysis revealed in total 70 loci that were associated with key parameters of depression-like and anxiety-related behaviours as well as with HPA axis reactivity in response to a psychological stressor. Some of the identified loci were associated with more than one key phenotype, reflecting the pluripotency of the loci detected. Associated loci were assessed regarding their functional relevance and some promising candidate regions were detected among protein-coding loci. Efforts are currently undertaken to define these regions more clearly and looking for gene dosage effects of CNVs. Additionally, as the identified loci require further validation to provide satisfactory evidence for their involvement in the genetic determination of depression and associated diseases, a second batch of 32 animals will be genotyped in order to confirm loci which were significantly associated in the first batch of mice.
Applying another powerful approach to pinpoint the genomic loci of quantitative traits such as HPA axis reactivity and emotionality, we plan to assess genome-wide multipoint oligogenic linkages by analysing a HRxLR-derived F2 intercross model, i.e. a population of freely segregating F2 mice (see Fig. 4). This approach, including principal component and quantitative trait locus (QTL) analyses, will support and maximize the output of our genetic studies, i.e. detecting genomic loci associated with the traits of interest in both, the unrelated CD-1 and the F2 populations derived from crossbreeding HR and LR animals of our stress reactivity mouse lines.
Additionally, we are currently planning to sequence the whole genome of our HR and LR mouse lines, looking for differences that might be involved in bringing about the clinically relevant endophenotypes of the model. For this endeavour, we are collaborating with scientists from the Wellcome Trust Sanger Institute in Cambridge, UK.
Taken together, these projects will yield powerful extensions of the results from our functional studies and could largely contribute to unravelling the molecular-genetic mechanisms underlying the behavioural, endocrine and neurobiological alterations observed in patients suffering from affective disorders.
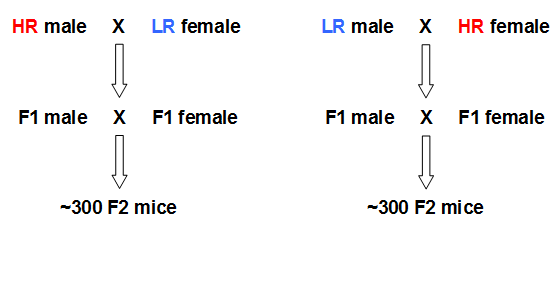
Breeding scheme for generating a freely segregating F2 population of mice derived from 16 HR and 16 LR mice (F0).

