Hauptinhalt
Topinformationen
Altered hippocampal energy metabolism in a mouse model of affective disorders: Insights from transcriptomic, proteomic and neurophysiological approaches
Gene and protein expression profiling experiments as well as neurophysiological assessments were conducted aiming to elucidate the relevance of differentially expressed genes, proteins and brain activity for the endophenotypes of the stress reactivity (SR) mouse model of affective disorders (HR/IR/LR lines).
Behavioral tests and magnetic resonance imaging approaches highlighted the hippocampus as a relevant brain area involved in the different phenotypes of the SR mouse lines. In vivo proton magnetic resonance spectroscopy was used to assess the levels of N-acetylaspartate (NAA) in the brain. NAA is a metabolite exclusively found in neurons and commonly used as a marker of neuronal integrity. Patients suffering from major depression and schizophrenia show decreased values of the NAA:creatine ratio in the hippocampus and prefrontal cortex (PFC). Interestingly, HR mice were found to have deficits in the performance of hippocampus- and PFC-dependent tests and showed decreased NAA levels in the right dorsal hippocampus and PFC. Moreover, the volume and functional activity of these brain areas in the SR mouse model were investigated using manganese-enhanced magnetic resonance imaging (MEMRI). The accumulation of manganese ions leads to signal enhancement in activated brain regions. The basal activity of the hippocampus was reduced in HR mice, although the hippocampal volume was not different between the three mouse lines. These results provided further support for the involvement of impaired hippocampal neural activity and a dysregulated HPA axis in the aetiology and pathophysiology of affective disorders.
The hippocampus is a known target of glucocorticoids and is subjected to structural and physiological alterations upon stress exposure. Furthermore, it is largely involved in the termination of the HPA axis stress response. Therefore, the following attempts to characterize the SR mouse model were focused on hippocampal tissue.
The transcriptome of HR vs. LR mice was investigated combining microarray-based and serial analysis of gene expression (SAGE) experiments. The detected pool of 981 differentially expressed genes in the hippocampus of HR vs. LR mice was further characterized via different bioinformatics tools. Cluster analyses were performed to systematically screen the set of regulated genes for functional enrichment. The database for annotation, visualization, and integrated discovery (DAVID), an integrative tool for the analysis of large sets of gene and protein lists, was employed. Regarding common biological processes, cellular components and molecular functions, Pathway Studio (8.0) was used to perform gene ontology analysis. Eight out of 14 significantly enriched clusters suggested mitochondrially active genes as the major player in differentiating between HR and LR mice on an expressional level. Importantly, the mitochondrial envelope, which harbors the respiratory chain proteins, was indicated in seven out of these eight clusters. Thus, our findings strongly suggest that, on a gene expression level, the differences in stress reactivity between HR and LR mice could be largely influenced by differences in mitochondrial function and, more precisely, in energy metabolism processes originating in the mitochondrial envelope.
Following this line of evidence, hippocampus tissue proteome studies were carried out. Proteomic alterations allow the identification of potential biomarkers and may be analyzed in the context of biochemical pathways in order to elucidate the biological mechanisms underlying molecular changes observed in affective disorders. Using two-dimensional gel electrophoresis and mass spectrometry, our analysis of HR and LR mice whole hippocampal lysates identified five differentially expressed proteins, three of them involved in metabolism/energy pathways.
In face of these results pointing to the mitochondria, we decided to further investigate specifically the mitochondrial proteome. Therefore, a comparative analysis of the hippocampal mitochondrial- and cytosolic-enriched proteomes from HR, IR and LR mice was conducted. Again using two-dimensional gel electrophoresis and mass spectrometry, ten proteins of the mitochondrial fraction and twelve of the cytosolic fraction were found to be differently expressed between HR, IR and LR mice. Not surprisingly, the majority of these proteins were involved in metabolism/energy pathways.
The results of our gene expression profiling in the SR mouse model allows to hypothesize that HR mice present a genetic predisposition to an elevated energy need in the brain. Presuming that the respiratory chain of HR mice complies with an elevated energy demand, an increased production of reactive oxygen species (ROS), the known side product of oxidative phosphorylation, would occur in HR compared to LR mice. One consequence of increased ROS levels is the induction of apoptosis via the intrinsic mitochondrial pathway, which is currently under investigation in our mouse lines.
Next steps of our approach will include the functional validation of candidate proteins using Western blotting, metabolite analysis and/or enzyme activity assays; ATP turnover differences; mutation frequencies between HR and LR mtDNA; activity state of antioxidant enzymes such as superoxide dismutase (SOD) and glutathione peroxidase. In addition, a time-dependent accumulation of pro-apoptotic signals might enable the detection of differing apoptosis rates in aged animals (see Figure).
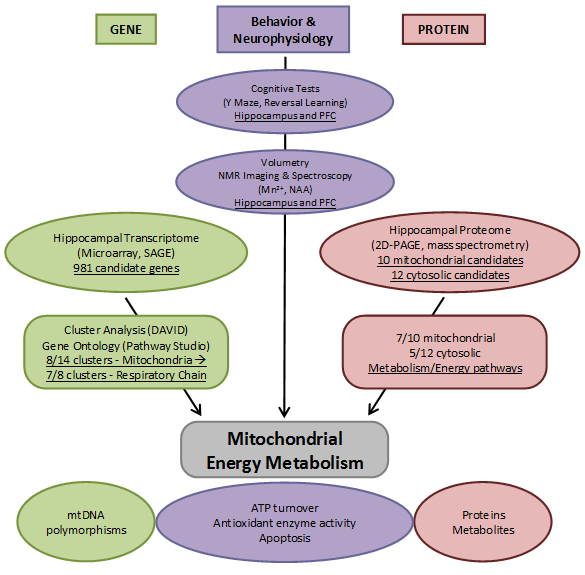
Overview of the results and approaches applied in the hippocampal transcriptome, proteome, and neurophysiology project addressing mitochondrial energy metabolism changes in the stress reactivity mouse model.

